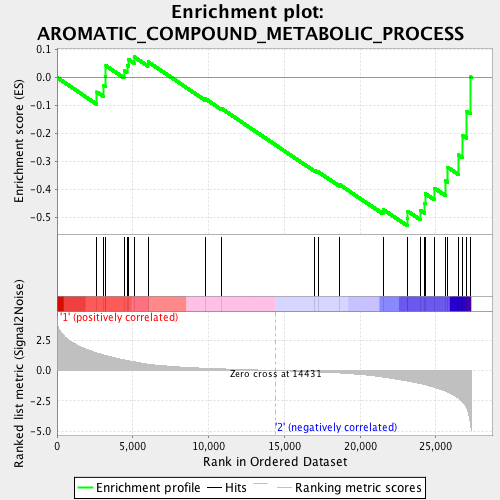
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | AROMATIC_COMPOUND_METABOLIC_PROCESS |
| Enrichment Score (ES) | -0.53157103 |
| Normalized Enrichment Score (NES) | -1.5789555 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.1865906 |
| FWER p-Value | 0.595 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MOCS2 | NA | MOCS2 Entrez, Source | molybdenum cofactor synthesis 2 | 2627 | 1.450 | -0.0517 | No |
| 2 | GSTZ1 | NA | GSTZ1 Entrez, Source | glutathione transferase zeta 1 (maleylacetoacetate isomerase) | 3039 | 1.296 | -0.0270 | No |
| 3 | SNCAIP | NA | SNCAIP Entrez, Source | synuclein, alpha interacting protein (synphilin) | 3198 | 1.241 | 0.0052 | No |
| 4 | ALDH6A1 | NA | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 3221 | 1.234 | 0.0423 | No |
| 5 | TGFB2 | NA | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 4414 | 0.880 | 0.0256 | No |
| 6 | PAICS | NA | PAICS Entrez, Source | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 4612 | 0.838 | 0.0441 | No |
| 7 | ALDH5A1 | NA | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 4715 | 0.806 | 0.0651 | No |
| 8 | HGD | NA | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 5078 | 0.726 | 0.0741 | No |
| 9 | PTS | NA | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 5997 | 0.527 | 0.0566 | No |
| 10 | HPD | NA | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 9774 | 0.189 | -0.0759 | No |
| 11 | DCT | NA | DCT Entrez, Source | dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) | 10873 | 0.138 | -0.1118 | No |
| 12 | TYR | NA | TYR Entrez, Source | tyrosinase (oculocutaneous albinism IA) | 17010 | -0.102 | -0.3335 | No |
| 13 | IDO1 | NA | 17227 | -0.111 | -0.3380 | No | ||
| 14 | CDA | NA | CDA Entrez, Source | cytidine deaminase | 18644 | -0.197 | -0.3838 | No |
| 15 | FTCD | NA | FTCD Entrez, Source | formiminotransferase cyclodeaminase | 21507 | -0.536 | -0.4722 | No |
| 16 | SULT1B1 | NA | SULT1B1 Entrez, Source | sulfotransferase family, cytosolic, 1B, member 1 | 23130 | -0.865 | -0.5051 | Yes |
| 17 | GMPS | NA | GMPS Entrez, Source | guanine monphosphate synthetase | 23147 | -0.868 | -0.4790 | Yes |
| 18 | MAT2B | NA | MAT2B Entrez, Source | methionine adenosyltransferase II, beta | 23983 | -1.084 | -0.4764 | Yes |
| 19 | MTHFD2 | NA | MTHFD2 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | 24253 | -1.166 | -0.4505 | Yes |
| 20 | QDPR | NA | QDPR Entrez, Source | quinoid dihydropteridine reductase | 24295 | -1.178 | -0.4159 | Yes |
| 21 | ASMTL | NA | ASMTL Entrez, Source | acetylserotonin O-methyltransferase-like | 24901 | -1.380 | -0.3957 | Yes |
| 22 | PRPS1 | NA | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 25617 | -1.696 | -0.3699 | Yes |
| 23 | ALDH1L1 | NA | ALDH1L1 Entrez, Source | aldehyde dehydrogenase 1 family, member L1 | 25762 | -1.774 | -0.3208 | Yes |
| 24 | SPR | NA | SPR Entrez, Source | sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) | 26483 | -2.280 | -0.2772 | Yes |
| 25 | HPRT1 | NA | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 26770 | -2.597 | -0.2081 | Yes |
| 26 | FAH | NA | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 27028 | -3.126 | -0.1216 | Yes |
| 27 | MOCOS | NA | MOCOS Entrez, Source | molybdenum cofactor sulfurase | 27272 | -4.325 | 0.0021 | Yes |