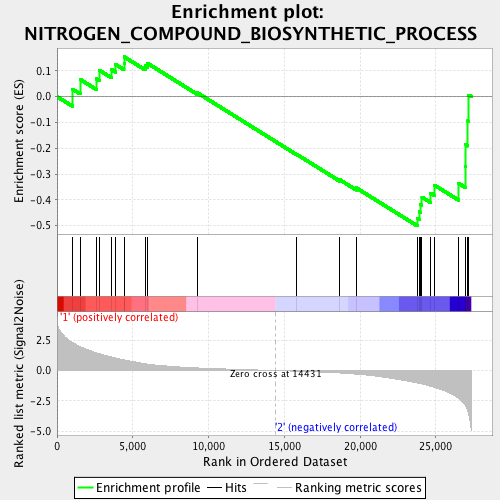
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | NITROGEN_COMPOUND_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | -0.50107336 |
| Normalized Enrichment Score (NES) | -1.5764476 |
| Nominal p-value | 0.008247423 |
| FDR q-value | 0.16411449 |
| FWER p-Value | 0.6 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BBOX1 | NA | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 1012 | 2.310 | 0.0291 | No |
| 2 | BCAT1 | NA | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 1516 | 1.968 | 0.0670 | No |
| 3 | CDO1 | NA | CDO1 Entrez, Source | cysteine dioxygenase, type I | 2583 | 1.467 | 0.0700 | No |
| 4 | AKT1 | NA | AKT1 Entrez, Source | v-akt murine thymoma viral oncogene homolog 1 | 2797 | 1.387 | 0.1019 | No |
| 5 | PLOD1 | NA | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 3593 | 1.121 | 0.1049 | No |
| 6 | SULT1A2 | NA | SULT1A2 Entrez, Source | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 | 3873 | 1.027 | 0.1241 | No |
| 7 | TGFB2 | NA | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 4414 | 0.880 | 0.1295 | No |
| 8 | DDAH2 | NA | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 4426 | 0.877 | 0.1542 | No |
| 9 | NQO1 | NA | NQO1 Entrez, Source | NAD(P)H dehydrogenase, quinone 1 | 5811 | 0.558 | 0.1195 | No |
| 10 | PAH | NA | PAH Entrez, Source | phenylalanine hydroxylase | 5957 | 0.535 | 0.1296 | No |
| 11 | HSP90AA1 | NA | HSP90AA1 Entrez, Source | heat shock protein 90kDa alpha (cytosolic), class A member 1 | 9268 | 0.215 | 0.0145 | No |
| 12 | SLC5A7 | NA | SLC5A7 Entrez, Source | solute carrier family 5 (choline transporter), member 7 | 15806 | -0.052 | -0.2234 | No |
| 13 | GLA | NA | GLA Entrez, Source | galactosidase, alpha | 18643 | -0.197 | -0.3216 | No |
| 14 | PRG3 | NA | PRG3 Entrez, Source | proteoglycan 3 | 19742 | -0.294 | -0.3534 | No |
| 15 | OAZ1 | NA | OAZ1 Entrez, Source | ornithine decarboxylase antizyme 1 | 23774 | -1.028 | -0.4716 | Yes |
| 16 | ETNK1 | NA | ETNK1 Entrez, Source | ethanolamine kinase 1 | 23889 | -1.061 | -0.4454 | Yes |
| 17 | MAT2B | NA | MAT2B Entrez, Source | methionine adenosyltransferase II, beta | 23983 | -1.084 | -0.4178 | Yes |
| 18 | ALDH18A1 | NA | ALDH18A1 Entrez, Source | aldehyde dehydrogenase 18 family, member A1 | 24075 | -1.110 | -0.3893 | Yes |
| 19 | PYCR1 | NA | PYCR1 Entrez, Source | pyrroline-5-carboxylate reductase 1 | 24654 | -1.294 | -0.3734 | Yes |
| 20 | ASMTL | NA | ASMTL Entrez, Source | acetylserotonin O-methyltransferase-like | 24901 | -1.380 | -0.3429 | Yes |
| 21 | SPR | NA | SPR Entrez, Source | sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) | 26483 | -2.280 | -0.3356 | Yes |
| 22 | HSP90AB1 | NA | HSP90AB1 Entrez, Source | heat shock protein 90kDa alpha (cytosolic), class B member 1 | 26948 | -2.904 | -0.2694 | Yes |
| 23 | GCH1 | NA | GCH1 Entrez, Source | GTP cyclohydrolase 1 (dopa-responsive dystonia) | 26961 | -2.929 | -0.1860 | Yes |
| 24 | EGFR | NA | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 27106 | -3.388 | -0.0942 | Yes |
| 25 | GCHFR | NA | GCHFR Entrez, Source | GTP cyclohydrolase I feedback regulator | 27159 | -3.574 | 0.0062 | Yes |