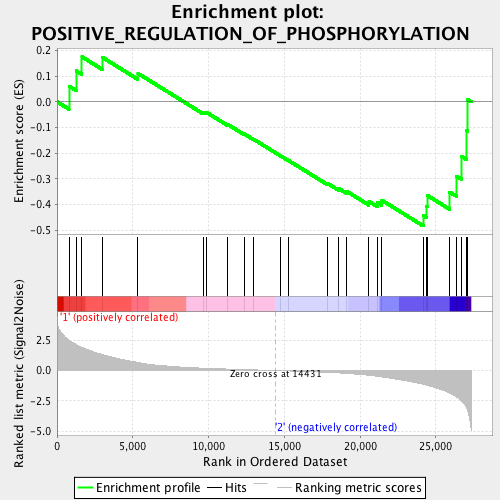
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | expression_mean_normalized_with_dChip_geneAvg_Mar1014_PNs.mainSample.cls #wt_versus_radiation.mainSample.cls #wt_versus_radiation_repos |
| Phenotype | mainSample.cls#wt_versus_radiation_repos |
| Upregulated in class | 2 |
| GeneSet | POSITIVE_REGULATION_OF_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.48323554 |
| Normalized Enrichment Score (NES) | -1.5312655 |
| Nominal p-value | 0.016460905 |
| FDR q-value | 0.18033975 |
| FWER p-Value | 0.714 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CCND2 | NA | CCND2 Entrez, Source | cyclin D2 | 785 | 2.489 | 0.0611 | No |
| 2 | EGF | NA | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 1262 | 2.127 | 0.1204 | No |
| 3 | CD81 | NA | CD81 Entrez, Source | CD81 molecule | 1640 | 1.900 | 0.1751 | No |
| 4 | TNK2 | NA | TNK2 Entrez, Source | tyrosine kinase, non-receptor, 2 | 2984 | 1.315 | 0.1734 | No |
| 5 | TDGF1 | NA | TDGF1 Entrez, Source | teratocarcinoma-derived growth factor 1 | 5335 | 0.652 | 0.1108 | No |
| 6 | BCL10 | NA | BCL10 Entrez, Source | B-cell CLL/lymphoma 10 | 9651 | 0.195 | -0.0402 | No |
| 7 | IL29 | NA | IL29 Entrez, Source | interleukin 29 (interferon, lambda 1) | 9835 | 0.186 | -0.0401 | No |
| 8 | ITLN1 | NA | ITLN1 Entrez, Source | intelectin 1 (galactofuranose binding) | 11219 | 0.122 | -0.0864 | No |
| 9 | CD80 | NA | CD80 Entrez, Source | CD80 molecule | 12340 | 0.077 | -0.1246 | No |
| 10 | CARD14 | NA | CARD14 Entrez, Source | caspase recruitment domain family, member 14 | 12968 | 0.054 | -0.1456 | No |
| 11 | IL3 | NA | IL3 Entrez, Source | interleukin 3 (colony-stimulating factor, multiple) | 14741 | -0.011 | -0.2101 | No |
| 12 | IL5 | NA | IL5 Entrez, Source | interleukin 5 (colony-stimulating factor, eosinophil) | 15268 | -0.032 | -0.2283 | No |
| 13 | IL20 | NA | IL20 Entrez, Source | interleukin 20 | 17826 | -0.143 | -0.3168 | No |
| 14 | HCLS1 | NA | HCLS1 Entrez, Source | hematopoietic cell-specific Lyn substrate 1 | 18598 | -0.193 | -0.3380 | No |
| 15 | EREG | NA | EREG Entrez, Source | epiregulin | 19078 | -0.233 | -0.3472 | No |
| 16 | IL12A | NA | IL12A Entrez, Source | interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) | 20579 | -0.390 | -0.3880 | No |
| 17 | ANG | NA | ANG Entrez, Source | angiogenin, ribonuclease, RNase A family, 5 | 21162 | -0.475 | -0.3922 | No |
| 18 | AKTIP | NA | 21430 | -0.521 | -0.3832 | No | ||
| 19 | IL31RA | NA | IL31RA Entrez, Source | interleukin 31 receptor A | 24163 | -1.138 | -0.4422 | Yes |
| 20 | GLMN | NA | GLMN Entrez, Source | glomulin, FKBP associated protein | 24365 | -1.200 | -0.4062 | Yes |
| 21 | BMP4 | NA | BMP4 Entrez, Source | bone morphogenetic protein 4 | 24438 | -1.222 | -0.3647 | Yes |
| 22 | CCND3 | NA | CCND3 Entrez, Source | cyclin D3 | 25911 | -1.862 | -0.3515 | Yes |
| 23 | CCND1 | NA | CCND1 Entrez, Source | cyclin D1 | 26352 | -2.173 | -0.2892 | Yes |
| 24 | CLCF1 | NA | CLCF1 Entrez, Source | cardiotrophin-like cytokine factor 1 | 26666 | -2.460 | -0.2119 | Yes |
| 25 | LYN | NA | LYN Entrez, Source | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 27040 | -3.155 | -0.1117 | Yes |
| 26 | EGFR | NA | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 27106 | -3.388 | 0.0082 | Yes |